Fundamentals of scRNASeq Analysis
2021-06-28 (updated: 2021-07-06)
Prerequisites
This is the teaching material for Session 2: Fundamentals of scRNASeq Analysis of 2021 Single Cell Workshop.
Install packages
Some packages used in this material can be installed from CRAN:
install.packages('Seurat')
install.packages("patchwork")
install.packages("dplyr")
install.packages("ggplot2")The R package hdf5r is also needed. For Windows user, you can simply install it from CRAN:
For Mac user:
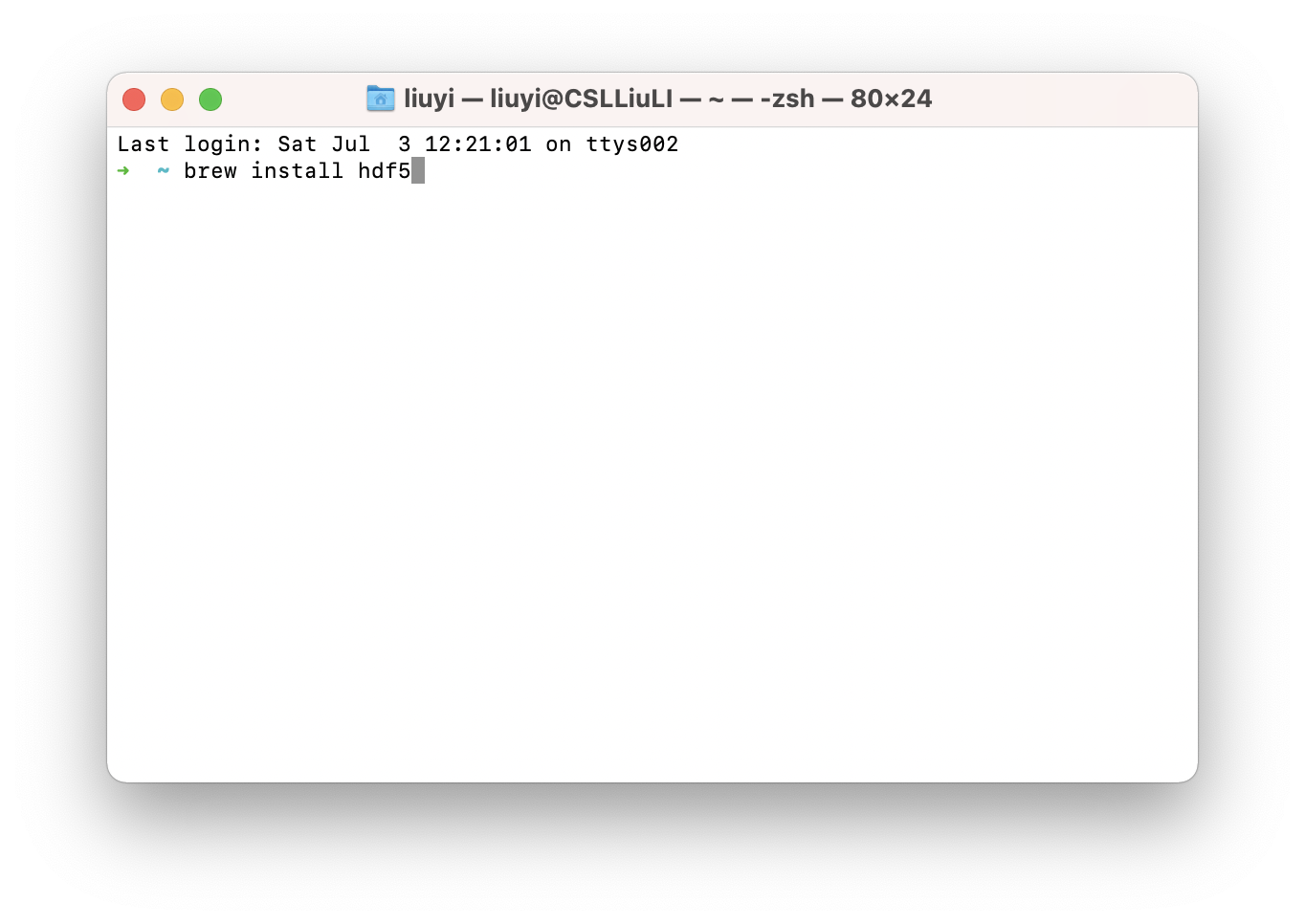
- open the
Terminalapplication and type followingtwocommands one by one:
/bin/bash -c "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/HEAD/install.sh)"
brew install hdf5
- If you got the error
command not found, you might need to add an extra command. In another word, instead of previous two command, you should type followingthreecommands one by one:
/bin/bash -c "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/HEAD/install.sh)"
echo "export PATH=/opt/homebrew/bin:$PATH" >> ~/.bash_profile && source ~/.bash_profile
brew install hdf5- open your R studio and install the R package
hdf5rfrom CRAN:
Set working directory
Setting a working directory first helps us read and save data in an easier way. To set a working directory:
create a folder named “R-workshop” in your preferred directory
create a “data” folder in the
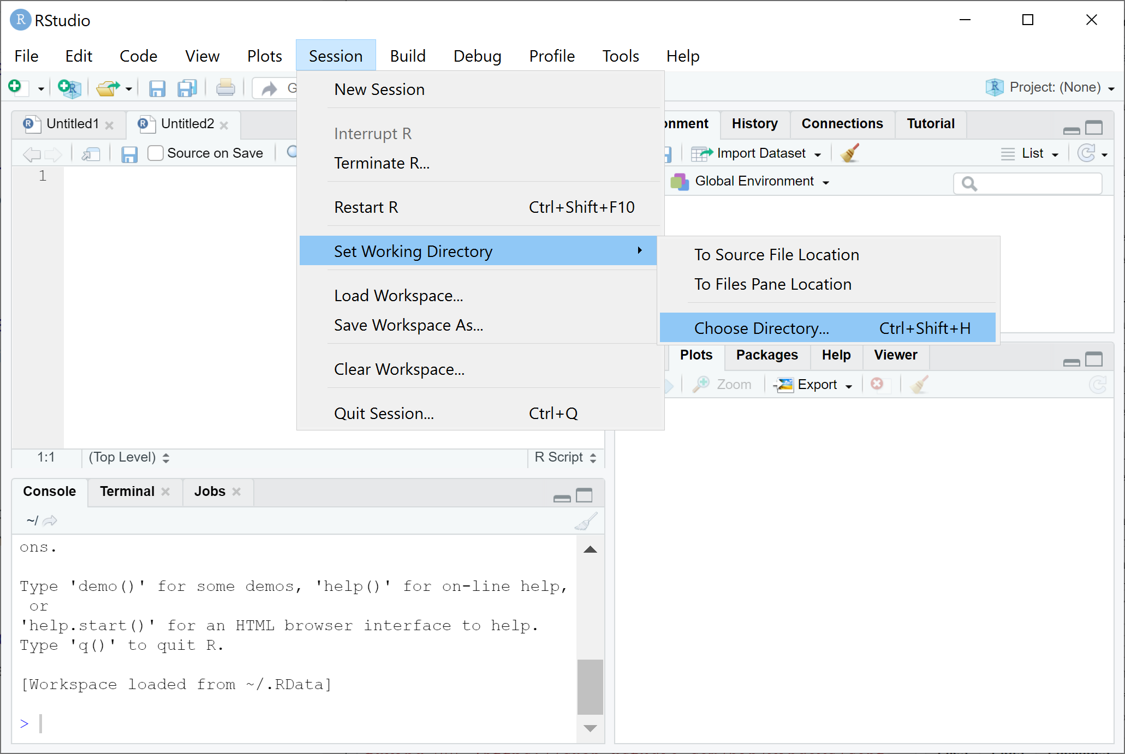
R-workshopfrom RStudio, use the menu to change your working directory under Session > Set Working Directory > Choose Directory
choose the directory to
R-workshop
Or you can type in the console:
setwd("/yourdirectory/R-workshop")For Windows, the command might look like :
setwd("c:/yourdirectory/R-workshop")Download data
Data used in this material is a 10k PBMC data getting from 10x Genomics website.
To prepare data for this workshop:
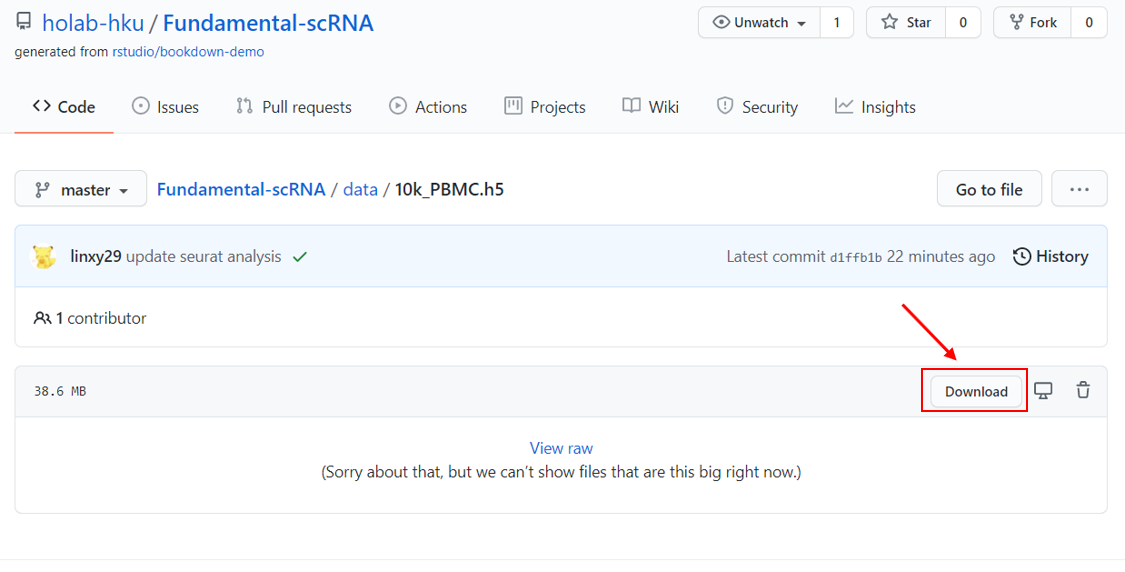
- download the
10k_PBMC.h5from: https://github.com/holab-hku/Fundamental-scRNA/blob/master/data/10k_PBMC.h5
put it in the
datafolder we create before.the
datafolder should be look like this:

Acknowledgement
Thanks everyone in Ho Lab who gives great comments/suggestions and contributes to this teaching material.
Other modules of this workshop
Module 1: Introduction to R by Dr Joshua Ho & lab Online tutorials
This is the Module 2: Fundamentals of scRNA-seq analysis by Dr Joshua Ho
Module 3: Cell clustering and annotation by Dr Asif Javed Online tutorials
Module 4.1: Cell interaction by Dr Asif Javed Online tutorials
Module 4.2 & Module 5: Cell trajectory inference & Cellular genetic analysis by Dr Yuanhua Huang Online tutorials